We study how different cell types in the human body are specified at the molecular level
Our research is animated by three major themes:
Transcription factor-chromatin interactions
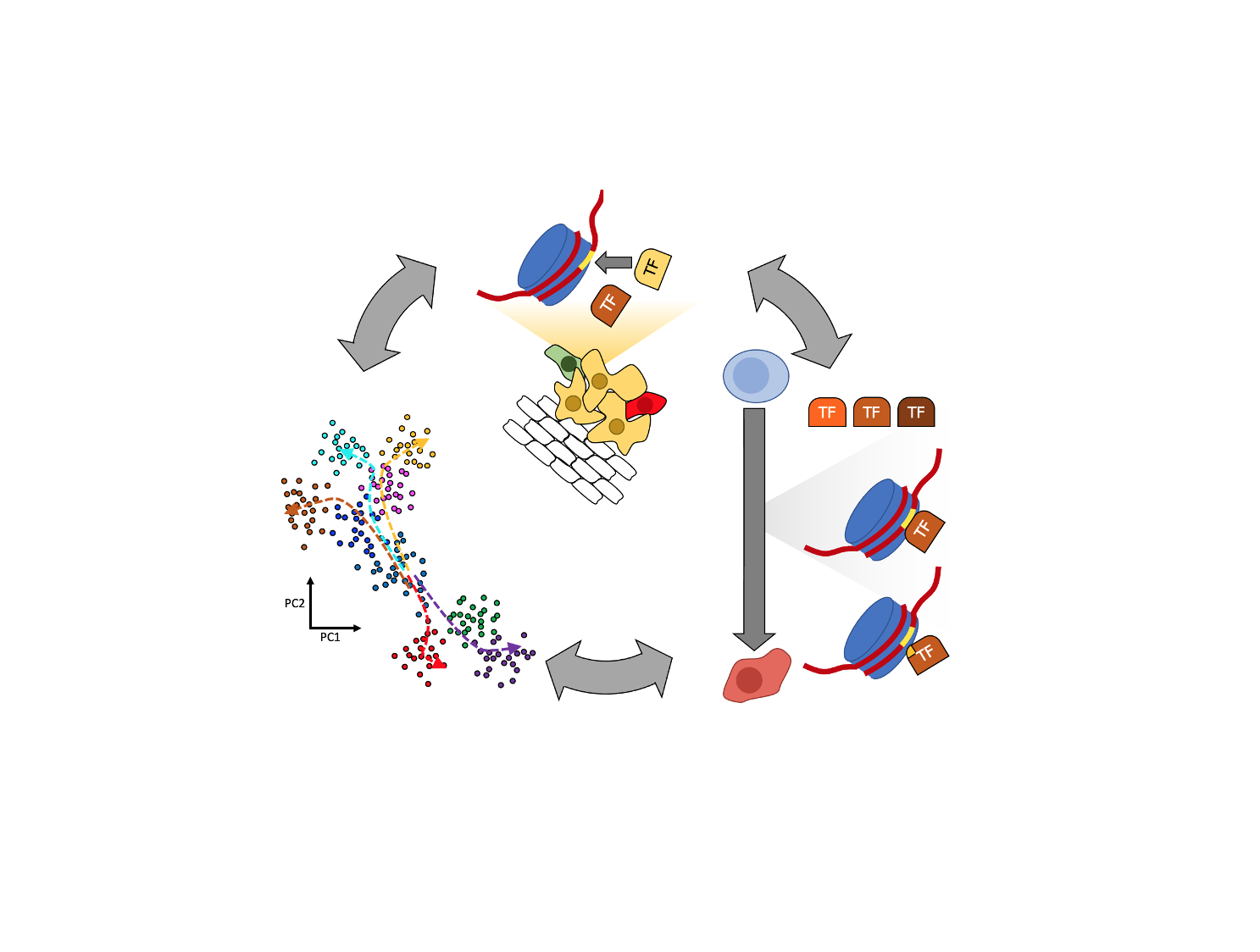
Mammalian genomes are decorated with millions of nucleosomes that each wrap ~150 base pairs of DNA in a stable configuration that enables efficient packaging into the confined nuclear space and denies access to the DNA by other nuclear factors. In order to specify cell fate, transcription factors bind to short DNA motif sequences in specific configurations throughout the genome to the exclusion of nucleosomes in order to activate the transcription of their constituent genes. A paradox therefore arises: How do transcription factors gain access to their motif targets in DNA when they are wrapped in nucleosomes to begin with? The answer to this question is fundamental to understanding how the fates of cells, our biological building blocks, are decided.
Cutting-edge genomics technology development
The specific positions where transcription factors, nucleosomes, and other proteins bind in the genome out of the billions of available DNA base pairs reflect how genes are regulated in the cell. High-throughput DNA sequencing techniques can be used to pinpoint these positions via technologies known collectively as “chromatin profiling” techniques. We develop new, cutting-edge chromatin profiling approaches deployed at scale and in individual cells to understand how transcription factors and nucleosomes are spatially distributed in different cell types, and how those spatial distributions change across time.
Chromatin dysregulation in disease
In cancer, particularly pediatric cancers that are unique in the relatively low number of somatic mutations they accumulate, the few genes that are mutated or otherwise genetically altered frequently encode proteins that contribute to chromatin structure and function. These chromatin-associated proteins employ disparate biological functions spanning global gene suppression to tissue-specific gene activation; they are ablated in a multitude of ways ranging from missense mutations to whole chromosomal rearrangements resulting novel gene fusions; and they manifest diseases as diverse as blood and brain cancers. We seek to better understand the ways in which such different cancers might be linked via their chromatin-centric pathogenesis.


